Annotations Exlorer
The annotations explorer allows users to explore the functional annotations of individual assemblies as well as search groups of assemblies.
Individual annotations can be accessed through links on the Homepage or via the
Search function. They can also be searched using the AlgaeDB ID code, or other identifying data, such as
NCBI assembly accession codes, or assembly names.
Annotations Explorer: Individual assemblies
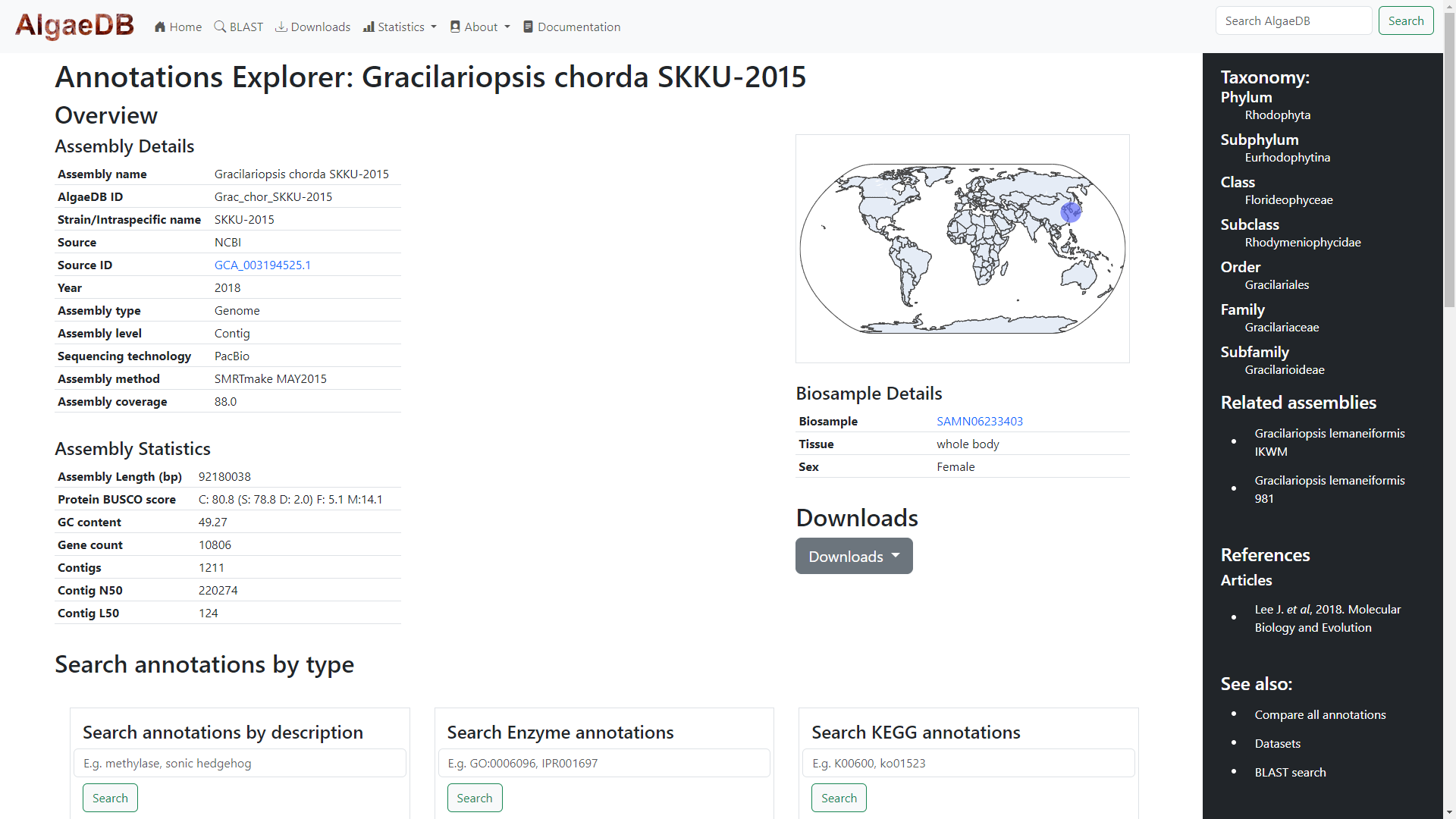
Annotation details of individual assemblies are viewed on dynamic pages. On the left, basic assembly details and statistics are shown, while
on the right sample details, taken from NCBI Biosample are shown.
These details are dynamic, and absent fields will not be shown. Above the Biosample table, a dynamic world map powered by Plotly shows the
location of the assembly sample. If the assembly has no available location data, then the map will state "No Location Data Available".
Below the Biosample details, a downloads button will be diplayed if downloads are available for the file. This activates a dropdown menu
of available files. Clicking on each item in the dropdown list will begin the download of the file.
Annotations can be explored using three search bars. Assemblies without functional annotations will not have these search functions available. These three bars can be used to
search annotations by name or description, Enzyme Code, Gene Ontology and InterPro annotations, or KEGG annotations, respectively.
These search bars can be used to query the functional annotation data, and function in the same way as the search page. The data that is
retrieved is the same between all three functions, but is displayed differently depending on the table searched.
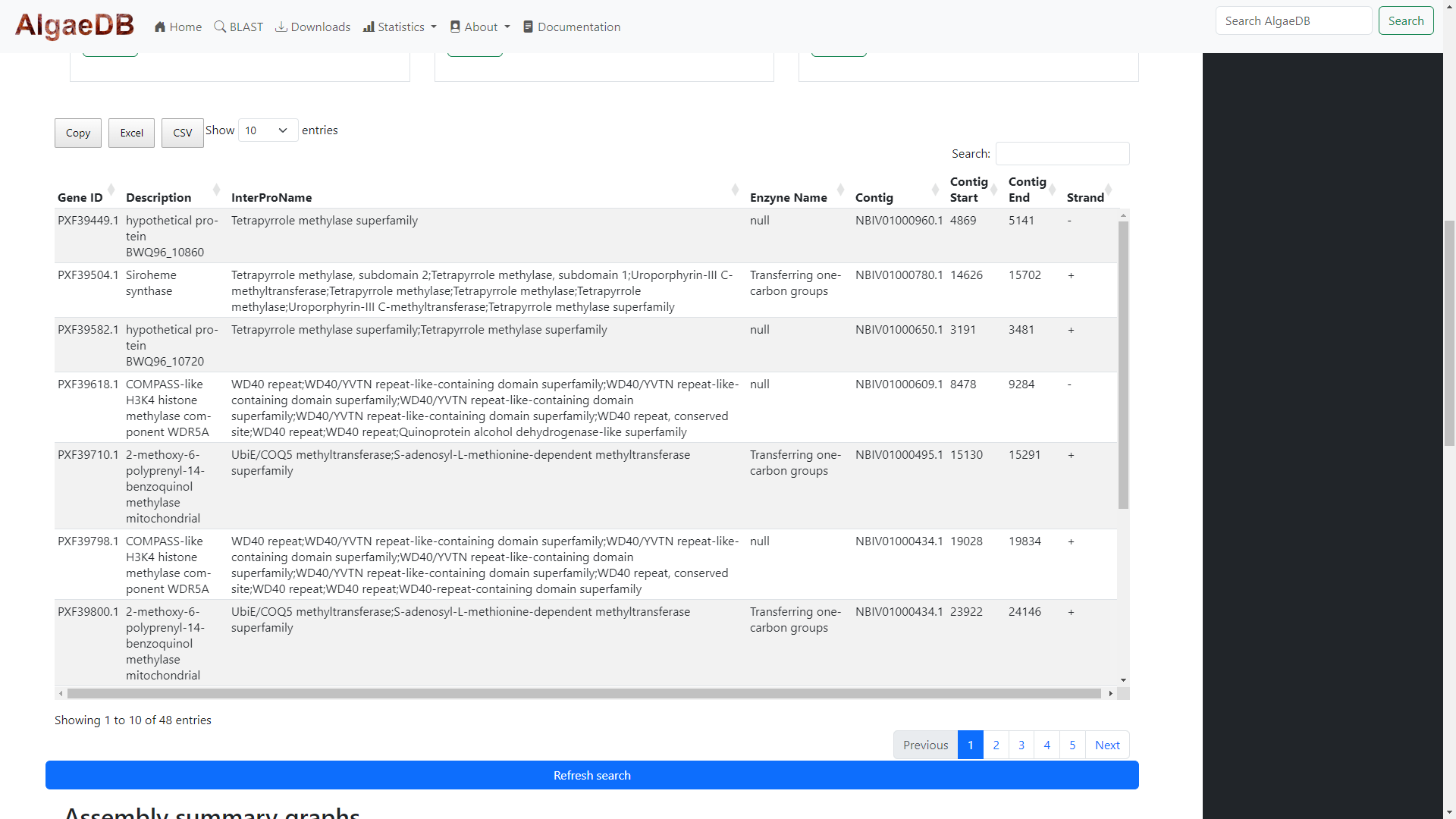
The three bars all search and retrieve the same data, but display different results depending on the search tab used. Results are displayed
on an interactive table powered by DataTables (for more information on dynamic tables, see the Dynamic tables section).